Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Scientific Reports volume 13, Article number: 22014 (2023 ) Cite this article Rna Prep Kit
Adonis mongolica is a threatened species that is endemic to Mongolia. It is a medicinal plant from the Adonis genus and has been used to treat heart diseases. However, the genomics and evolution of this species have not been thoroughly studied. We sequenced the first complete plastome of A. mongolica and compared it with ten Adonideae species to describe the plastome structure and infer phylogenetic relationships. The complete plastome of A. mongolica was 157,521 bp long and had a typical quadripartite structure with numerous divergent regions. The plastomes of Adonideae had relatively constant genome structures and sizes, except for those of Adonis. The plastome structure was consistent across Adonis. We identified a 44.8 kb large-scale inversion within the large single-copy region and rpl32 gene loss in the Adonis plastomes compared to other members of the Adonideae tribe. Additionally, Adonis had a smaller plastome size (156,917–157,603 bp) than the other genera within the tribe (159,666–160,940 bp), which was attributed to deletions of intergenic regions and partial and complete gene losses. These results suggested that an intramolecular mutation occurred in the ancestor of the Adonis genus. Based on the phylogenetic results, Adonis separated earlier than the other genera within the Adonideae tribe. The genome structures and divergences of specific regions in the Adonis genus were unique to the Adonideae tribe. This study provides fundamental knowledge for further genomic research in Mongolia and a better understanding of the evolutionary history of endemic plants.
Morten E. Allentoft, Martin Sikora, ... Eske Willerslev
Morten E. Allentoft, Martin Sikora, ... Eske Willerslev
Daqi Yu, Yandong Ren, … Juan Pascual-Anaya
Adonis L., comprising 26–38 species, belongs to the Ranunculaceae family1,2,3. The higher classification of Adonis is the Adonideae tribe4,5. This genus is commonly distributed in southwestern Asia, Europe, Northern Africa, and the Mediterranean region1,6. The general morphology of Adonis is characterized by therophytes or perennials, compound leaves, 1–2 palmately or 2–3 pinnately dissected terminal inflorescences, and white, red, and yellow flowers6,7. The base number of chromosomes in the genus is x = 8. In perennial species, the somatic number of chromosomes is 2n = 16, and polyploid races with 2n = 24 and 2n = 32 have been identified8,9. Adonis is divided into two subgenera, Adonanthe and Adonis, which are perennial and annual species, respectively.
Adonis mongolica Simonovich is an important medicinal plant that is endemic to Mongolia10,11. Since 2000, it has been identified in only two locations in the country, which is of great concern12. The general morphology of A. mongolica is a plant height of 10–20 cm, with numerous white flowers and violet sepals (Fig. 1). A. mongolica inhabits a specific environment compared with other Adonis; however, previous studies on A. mongolica have primarily focused on its chemical components and medical usage13,14,15, in addition to its distribution and conservation12,16. However, the genetic background of A. mongolica, along with its phylogenetic relationships and classification, has not been determined. Similar to other Adonis species, A. mongolica has been used to treat heart failure with symptoms of tachycardia and edema13. Currently, A. mongolica is critically endangered globally because of overexploitation and loss of habitat16. Therefore, implementing programs for protecting and genetic breeding of threatened species is critical. Most Adonis species grow where topographical conditions allow for a water supply17, but A. volgensis, A. villosa, and A. mongolica grow mostly on drier montane and forb steppes, and these species show evidence of ecological vicariance18,19,20,21. The evolution of A. mongolica was most likely accompanied by the alteration and extension of its ecological constitution.
Photographs of wild Adonis mongolica of (a) general habitat, (b) mature flower, natural habitat, (c) flower, front view, (d) flower, dorsal view, (e) leaves, (f) aggregated fruit. Photographs by B. Oyuntsetseg (a) and Ch. Javzandolgor (b–f).
Over the past few decades, several researchers have studied the whole chloroplast genome (plastome) sequences of plants worldwide. This is because using the plastome sequences to resolve relationships between vascular plants at higher taxonomic levels is more successful than using short-sequence fragments22,23. Furthermore, in most angiosperm species, plastome inheritance is uniparental and highly conserved and provides important phylogenetic data24. The first detailed plastome study of Adonis in 1998 described the restriction site maps for the two species25. Currently, the plastomes of four Adonis species are available in the NCBI GenBank database26,27,28, accounting for only 2% of the total species diversity. According to previous plastome studies, the plastomes of the genus Adonis have large inversions and gene transpositions25,27, and the genus belongs to the Adonideae tribe along with Calathodes, Megaleranthis, and Trollius26,27,28. The Adonideae tribe was derived from its ancestor around 25.5 Mya28. In addition, phylogenetic relationships within the Adonis genus have been evaluated using partial barcoding genetic markers such as nuclear internal transcribed spacers (ITS)6,29,30,31, trnL-F29, and random amplified polymorphic DNA (RAPD) markers30. However, these studies mostly used several species in sect. Adonanthe, and the entire Adonis genus has not been studied. Therefore, our study provides fundamental phylogenomic data for Adonis and lays the foundation for future phylogenetic studies of Adonideae.
This study aimed to characterize the plastome of A. mongolica and identify genetically variable regions through comparison with closely related species within the Adonideae tribe. The plastome results provide useful information for future studies, such as evolutionary analyses and safe medical applications of A. mongolica.
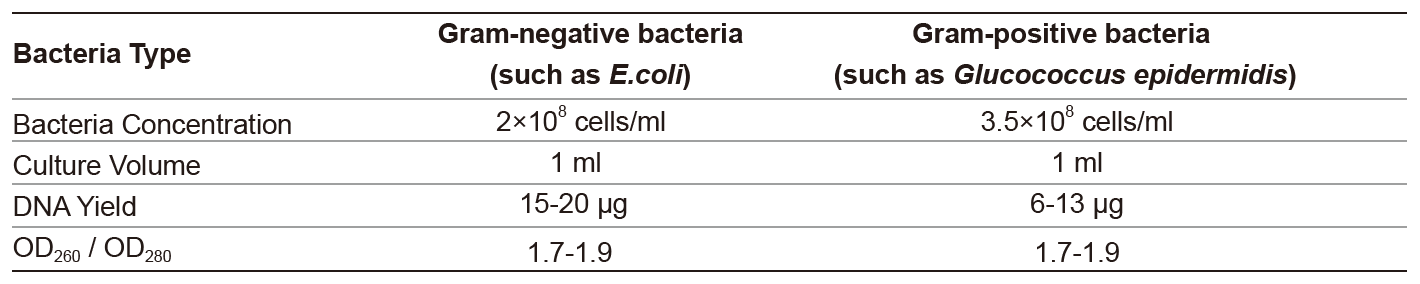
The plastome of A. mongolica was sequenced for the first time. A total of 8 Gb paired-end (150 bp) clean reads were generated. After trimming and normalization, a list of 22,993,276 paired reads was obtained for de novo assembly (Table S1). De novo assembly generated a single contig, 157,521 bp in length. The plastome of A. mongolica contained a large single-copy (LSC; 86,651 bp), a pair of inverted repeats (IR; 26,332 bp), and a small single-copy (SSC; 18,206 bp). The total GC content in A. mongolica was 37.9%. Particularly, the GC contents in the IR, LSC, and SSC regions were 43, 36.1, and 31.4%, respectively (Table 1), indicating that the IR region had a higher GC content than the SSC and LSC regions. Adonis mongolica contained 113 unique genes. The numbers of rRNA, tRNA, and protein-coding genes in the plastome were 4, 30, and 79, respectively (Fig. 2 and Table S2). Overall, the protein-coding sequences (CDS) were 78,470 bp long, whereas the non-coding sequences (including the intergenic spacer [IGS] and introns) were 79,051 bp long.
Circular gene map of the complete chloroplast genome of Adonis mongolica. Genes drawn inside the circle are transcribed clockwise, and those outside, counterclockwise. The darker gray in the inner circle represents GC content and light gray represents AT content. Intron containing genes are marked with a star.
The plastome sequences of 11 Adonideae species (A. mongolica, A. amurensis Regel & Radde, A. pseudoamurensis W.T.Wang, A. sutchuenensis Franch., A. coerulea Maxim., Calathodes oxycarpa Sprague, Megaleranthis saniculifolia Ohwi, Trollius macropetalus (Regel) F. Schmidt ex W.T.Wang, T. chinensis Bunge, T. farreri Stapf, and T. ranunculoides (Sm.) Steam) were analyzed in this study (Table S3). The complete circular plastomes were 156,917–160,191 bp in length and included an LSC of 86,218–88,944 bp, an SSC of 18,067–18,532 bp, and a pair of IR regions of 26,087–26,632 bp (Table 1 and Table S4). The overall GC content was 37.9–38.1% and was slightly higher in the IR than in the LSC and SSC regions. The Adonideae plastomes contained 131 genes, including 80 protein-coding, 30 tRNA, and 4 ribosomal RNA genes (Fig. S1). Six protein-coding, seven tRNA, and four rRNA genes were duplicated within the IR regions. The complete plastome harbored 18 intron-containing genes, 16 of which had a single intron and two of which had two introns (Tables S5 and S6). Three intron-containing genes were found to be duplicated in the IR regions. The rps12 gene was a trans-spliced gene with the 5′-end located in the LSC and the 3′-end located in an IR region.
Analysis of codon usage and anticodon recognition patterns revealed 26,210–26,427 codons, of which leucine, serine, and isoleucine were the most abundant (Fig. S2). To identify codon patterns, we analyzed codon distribution in 11 plastomes (Fig. S3). Codons with A or T at the third position had a strong codon bias. Most Adonideae displayed a comparable pattern with high RSCU values for arginine (AGA), leucine (TTA), and alanine (GCT).
We investigated the repeat sequences of Adonideae species to determine their characteristics and proportions of repeat sequences within the tribe. Adonideae species had a similar number of forward, reverse, palindromic, and complementary repeats (Fig. 3a). For single-sequence repeats (SSRs), mononucleotide motifs were the most abundant in all species, followed by dinucleotide motif repeats (Fig. 3b). A total of 53–69 SSRs were identified, mostly in the LSC region and particularly within the IGS region. Most of the tandem repeats were located in the IGS region (Fig. 3c). We found 23–58 tandem repeats that were generally 9–68 bp long (Fig. 3d). Among the tandem repeats, three regions (rps16-ndhJ, ndhF-ccsA, ndhG-ndhI) in the IGS and the ycf2 gene in the LSC region were common among the five Adonis plastomes. The number of repeat sequences was the highest in T. macropetalus (Fig. 3e). However, the tandem repeats were shorter than the other repeat sequences, and this pattern was similar in all the Adonideae examined (Fig. 3f).
The distribution of repeat sequences in of tribe Adonideae plastomes. (a) The number of repeat types in Adonideae plastome. (b) The number of simple sequence repeats (SSR) motifs and the distribution of SSRs in the genome regions. (c) Distribution of tandem repeats in intergenic region (IGS), intron, and exon. (d) Total number of tandem repeats. (e) Distribution of repeats in genomic regions. (f) Percentage of repeats in genomic regions.
The genome structure, gene order, and boundaries between IRs and single-copy regions were similar among Adonideae plastomes; however, genus-specific rearrangements in Adonis plastomes were revealed. Collinearity in gene placement among the 11 available Adonideae plastomes was assessed using AliTV (Fig. 4 and Fig. S4). From the trnT-UGU to the rps16 genes, a 35-gene inversion (21 protein-coding genes and 14 tRNAs), 44.8 kb in length, evolved in Adonis. Furthermore, two deletion sites (844 and 524 bp) in the intergenic regions were found within the inversion region of the Adonis plastomes. This inversion was not detected in other available species within the tribe. In Adonideae, IR lengths ranged from 26,087 to 26,632 bp, but the borders between the IR regions and the two single-copy regions (LSC and SSC) were similar. The rps19 and ycf1 genes spanned the LSC/IRb and SSC/IRa junctions, respectively (Fig. S5). Only IR contraction was observed in A. coerulea because the rps19 gene was located entirely in the LSC region.
The complete plastome structure of Adonideae species. Linear maps were built using the AliTV visualization software, based on whole genome alignment. Both panels depict pairwise comparisons, expressed as percentage of nucleotide similarity, that connect different homologous genomic regions. Genomes are completed in full and pictured in purple.
The sequence identities of the 11 Adonideae were analyzed using mVISTA, with the A. mongolica plastome serving as a reference. We replaced the inversion of Adonis to calculate the sequence identity of each gene. As expected, the genic regions were more conserved than the intergenic regions when comparing the ten species (Fig. S6). This pair of IR regions was highly conserved, followed by the LSC and SSC regions. We analyzed the genetic divergence of genes and intergenic regions within Adonideae plastomes, and genus-related mutations were detected using pairwise comparisons. Overall, Adonideae plastomes had an average genetic diversity (Pi) value of 0.018. The highest Pi values of the IGS regions were observed for trnK-rps16 (0.102) in SSC (Fig. 5). The ycf1, rps16, ndhF, and matK genes are variable within the Adonideae species. Species-specific mutations of Adonis were detected using pairwise alignment. The highest Pi value was detected for psbT-psbN (0.104) in A. mongolica and A. coerulea (Fig. S7). In addition, the ndhF-trnL IGS showed considerable length variation in Adonideae because the rpl32 gene was completely lost in Adonis (Fig. S8).
Comparison of the nucleotide diversity (Pi) values of intergenic regions among Adonideae. The mean Pi value of intergenic regions within the tribe Adonideae and the genera Adonis and Trollius is indicated by green, blue and orange lines, respectively.
Two datasets were compiled to verify the phylogenetic relationships among members of the Adonideae tribe. The first dataset consisted of 80 conserved protein-coding sequences of 11 Adonideae species, obtained by aligning 87,075 bp, of which 82,024 bp (94.2%) were constant and 5051 bp (5.8%) were parsimony-informative. The second dataset contained the complete plastomes of 17 Ranunculaceae species. Due to the relatively high variation within the intronic and intergenic regions, the matrix contained 200,666 nucleotide sites, of which 152,797 bp (76.1%) were constant and 37,887 bp (18.9%) were parsimony-informative among Ranunculaceae. The Aconitum kusnezoffii Rchb. plastome served as the outgroup for both datasets. The Adonis species were distinct from other genera in the Adonideae tribe and were supported by strong bootstrap values and posterior probabilities (PPs) for the CDS and whole plastome datasets (Fig. 6 and Fig. S9). Adonis mongolica clustered with A. coerulea and A. sutchuenensis, whereas A. amurensis and A. pseudoamurensis were positioned as sister taxa in this cluster. However, the relationship between A. mongolica, A. coerulea, and A. sutchuenensis was differently positioned in the two datasets: in the whole plastome dataset, A. mongolica and A. coerulea were more closely related than A. sutchuenensis (Fig. 6); while, in the CDSs dataset, A. coerulea and A. sutchuenensis were more closely related than A. mongolica (Fig. S9).
Phylogenetic tree from Ranunculaceae species based on the whole plastomes using Maximum parsimony (MP), Bayesian inference (BI), and Maximum likelihood (ML). The MP topology is indicated with BI probabilities and ML bootstraps at each branch; maximum support values are not indicated (MP 100, PP 1, and ML 100). The plastome completed in this study is indicated with bold font.
Plastomes have been widely used as models to elucidate patterns of genetic variation in space and time, as well as micro- and macro-evolutionary events across all plant lineages32. This is because the plastome is highly conserved, with evolutionary hotspots, such as gene insertions and deletions, IR contractions and expansions, inversions, and various repeat sequences33. We sequenced and characterized the plastome of A. mongolica and conducted comparative studies that included ten other species of the Adonideae tribe to identify genetic variations that could explain the evolutionary changes. The complete plastome of A. mongolica showed a typical quadripartite structure with one LSC, one SSC, and two IR regions, which was a highly conserved pattern. However, unique structural rearrangements were detected in the newly sequenced A. mongolica, as with other species of Adonis. Multiple inversions and transpositions have been detected in the LSC regions of Ranunculaceae plastomes27,34. The Adonis genus plastome includes a 44.8 kb inversion in the LSC region25,27,28. Inversions arise when two breaks occur within a DNA segment, and the segment between the two breaks inserts itself in the opposite direction, causing a 180° inversion relative to the adjacent region. Inversion breakpoints are frequently found in regions with repetitive nucleotides, and these regions may be reused in future inversions35. Repeated sequences have been discovered at the endpoints of several land plant plastome inversions, as well as in tRNA genes36. In the Adonis genus, the inversion reversed the order of genes between rps16 and trnT-UGU, placing trnK-UUU adjacent to trnT-UGU and rps16 adjacent to trnL-UAA. This inversion occurred between two tRNA genes; however, there was no repeat region near the endpoint of the inversion. This might have been caused by random changes in the Adonis plastome. Furthermore, large-scale inversion occurred in the ancestor of Adonis after being derived from the Adonideae tribe because this type of inversion was not observed in any other genus within the tribe. Plastome sequences have been used by many researchers to determine phylogenetic relationships at various taxonomic levels37. Early molecular phylogenetic studies have revealed that Adonis is related to Trollius38. Based on floral development and phyllotaxis, Adonis was assigned to the Adonideae tribe and is closely related to the genus Calathodes5. In addition, the phylogenetic relationship of Ranunculaceae was revised using plastome analysis, revealing that Adonis belonged to the Adonideae tribe27,28,39. Our phylogenetic study revealed the same topology as previous studies; the Adonideae tribe formed a monophyletic clade, which includes Adonis, Calathodes, Megaleranthis, and Trollius based on whole plastome and protein-coding genes. Within this tribe, the Adonis genus formed a distinct monophyletic group (Fig. 6).
The plastome size of the Adonideae species investigated here varied slightly, with that of the Adonis genus being shorter (156,917–157,603 bp) than those of the other Adonideae species (159,666–160,940 bp). Adonis plastome size diversity was detected in LSC and IR. The IR contraction occurred due to a partial deletion (200 bp) of the ycf2 gene, reduction of the LSC region due to a ~ 2 kb nucleotide loss during large-scale inversion, and intergenic deletions in rps16-trnQ-UUG and psbM-trnD-GUC regions. The ycf2 gene is the largest plastid gene reported in angiosperms. The function of ycf2 is largely unknown; however, it does not appear to be related to photosynthesis40. The ycf2 gene within Ranunculaceae species shows genetic divergence with many repeat events and deletions41,42. The ycf2 gene could be an alternative to comprehensive genes for studying the relationships between Ranunculaceae species. More than 69 SSRs were identified in A. mongolica, and most repeats were in the ycf2 gene. This provides a rare opportunity to study the population genetics of this threatened plant, which is endemic to Mongolia. In addition, length variation was detected in the ndhF-trnL intergenic region within Adonideae. This spacer includes the gene rpl32, which produces the ribosomal protein L3243; however, the gene has been completely deleted in the Adonis genus27,34,39,44. This spacer within the Ranunculaceae plastome is either a pseudogene or absent and has a wide range of lengths with a 1.6 to 5.5-fold reduction compared to that of the full-length IGS of rpl3245. Due to various events of complete loss, this IGS region in Adonis is nearly 0.8 times shorter than that in other Adonideae genera. Transfer of rpl32 from the plastome to the nucleus has been observed in Euphorbiaceae46, Ranunculaceae27,45, Rhizophoraceae47, and Salicaceae48; thus, the rpl32 may have been transferred to the nucleus or subjected to complete loss in the Adonis plastome. Furthermore, some coding regions had significantly high diversity, such as the ycf1, rps16, and matK genes (Fig. 5). These highly variable regions may be useful as specific DNA barcodes for species-level identification, as well as for providing genetic markers for resolving relationships among the Adonideae.
Taxonomic studies of the Adonis genus have not been well revised; the species of sect. Adonanthe were mostly studied, including the nuclear ITS region6,29,30 and the trnL-F29, atpB, and rbcL38 regions of the plastome. In the present study, we estimated the genetic diversity of the plastomes of five species of sect. Adonanthe. The ccsA-ndhD, ndhD-ndhI, and trnD-trnY regions showed different haplotypes for each species (Fig. S7). These regions can be used to identify species within the Adonis genus. Among the modern barcoding markers, trnH-psbA (0.023) was highly variable for Adonis, whereas the previously used trnL-F (0.006), atpB (0.005), and rbcL (0.004) markers were relatively less variable. Based on our observations, partial genetic markers of the plastome are unsuitable for identifying species within the Adonis genus. Based on whole plastome sequences, A. mongolica clustered with A. coerulea and belonged to subg. Adonanthe sect. Adonanthe ser. Coerulea. The results revealed the morphological characteristics of petiolate leaves, rhizomes, trichomes on achenes, and white or purple petals49. The species of ser. Coerulea has numerous mixed light-purple and white flowers, whereas A. mongolica and A. davidii have only white flowers7,18. Interestingly, A. mongolica was a sister taxon to the cluster containing A. coerulea and A. sutchuenensis based on protein-coding genes. Specific mutations may have occurred in protein-coding genes of A. mongolica. Many studies have revealed that mutations in genes corresponding to flower color can affect adaptive success50,51. Future studies on the gene expression of flower color will be valuable for a better understanding of the adaptation and evolution of the Adonis genus.
The results of the present study expand the genetic information on medicinal plants endemic to Mongolia. In particular, A. mongolica has numerous genetic divergence regions; Adonis members show a genus-specific 44.8 kb inversion, several partial deletions, and complete gene loss. These changes have not been detected in other genera within the tribe; thus, they occurred in the ancestor of Adonis after its separation from the Adonideae tribe. This may have been caused by nonrandom recombination associated with climate change, making it an interesting topic for future evolutionary investigation.
The plastome of A. mongolica was sequenced and annotated for the first time. It included a highly conserved gene order, identical to that of the Adonis species, at 157 kb, and has a median plastome size for photosynthetic land plants. We identified important genetic resources for sequence divergence and phylogenetic inference of A. mongolica, providing a rare opportunity to study the genetic background of this threatened plant endemic to Mongolia. Comparative genomics indicated that Adonis plastomes were relatively conserved but included several genus-specific rearrangements and highly divergent locations. For instance, we identified a uniquely large inversion in the LSC region, complete gene loss from the SSC region, and some genes and intergenic regions with high sequence diversity. This study provides valuable genetic information to understand the evolutionary relationships of the genus Adonis within the Adonideae tribe.
The plant material used in this study was collected from natural populations in Argalant Soum, Tuv province (N47.887992 and E106.41658), Mongolia, and a voucher specimen (UBU0032866) was deposited in the herbarium of the National University of Mongolia, Mongolia. Total DNA was extracted from the tested species by using a modified CTAB method52. The library was prepared from total genomic DNA using the TruSeq DNA Nano Kit on the NextSeq 500 platform (Illumina, San Diego, CA, USA), following the manufacturer’s protocol. Trimmomatic v. 0.3653 was used to remove adapter sequences and low-quality reads to reduce biases in the analysis. The total number of bases, reads, GC (%), Q20 (%), and Q30 (%) for A. mongolica samples were calculated after filtering. A base quality plot generated using FastQC v. 0.11.554 was used to evaluate the overall quality of the data. This plot shows the range of quality values for each cycle. De novo assembly was performed with various k-mers using NOVOplasty55. The best k-mer was selected based on the assembly results, including the number of contigs, total contig length, and N50. Read sequences produced using the Illumina platform for plastome assembly were mapped to validate the assembly results, and the depth, coverage, and insert size were calculated. The raw data reads were mapped to an assembly result to identify the insert size of the raw data and the number of reads used in the assembly. The appropriate statistics were calculated after mapping. After the complete or draft plastome was assembled, BLAST analysis was performed to identify the species with which each contig (or scaffold) showed similarity.
We used REPuter to identify forward, reverse, palindromic, and complementary repeats with a minimum length of 20 bp, 90% identity, and a Hamming distance of 356. SSRs were detected using MISA57 with the minimum number of repeat parameters set to 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. Tandem repeats ≥ 20 bp were identified using the Tandem repeats finder58 with a minimum alignment score of 50 and a maximum period size of 500; the identity of repeats was set to ≥ 90%.
Plastome annotation of A. mongolica was performed using GeSeq59 to predict the location, whereas BLAST60 was used to determine the function of and identify the assembled sequences against nucleotide and protein sequence databases. The complete plastome sequence of A. mongolica has been submitted to GenBank (accession number: OQ569932). A total of 11 samples, representing five species from the genus Adonis, including A. mongolica, four species from the genus Trollius, and one species each from the genera Megaleranthis and Calathodes, were analyzed in this study. OrganellarGenomeDraw61 was used to generate circular and linear plastome maps. The GC content and RSCU of the ten plastomes were analyzed using MEGA11 software62. The codon usage distribution of Adonideae plastomes was visualized using the Heatmapper tool with average linkage clustering and Euclidean distance measurement methods63. An RSCU < 1.00 indicated a codon that was used less frequently than expected, whereas an RSCU > 1.00 indicated a codon that was used more frequently than expected. Pairwise whole plastome alignments of the Adonideae plastomes were visualized using AliTV software64 and MAUVE v2.3.165. The SC/IR boundary shifts at four junctions (LSC/IRa, IRa/SSC, SSC/IRb, and IRb/LSC) of the sample plastomes were compared using IRscope66. The mVISTA program67 in Shuffle-LAGAN mode68 was used to detect species-specific genetic variation by comparing Adonideae plastomes with A. mongolica as a reference. DnaSP version 669 was used to calculate nucleotide variability (Pi) among plastomes. The CDS, intron, and IGS regions were analyzed separately to confirm the exact genetic variants. Statistical analysis was performed using XLSTAT70 to test whether gene loss was related to IGS length.
We constructed two datasets for phylogenetic analysis: CDSs of Adonideae species and complete plastomes of Ranunculaceae species. Complete plastomes and CDSs were individually aligned using MAFFT ver. 7.38871. Alignment datasets were filtered to remove ambiguously aligned regions using GBlock ver. 0.91.172. The best-fitting model for nucleotide substitutions was determined using the Akaike information criterion in jModelTest v2.1.1073, and the GTR + I + G model was selected for maximum likelihood (ML) analysis. The GTR + G model was selected for the Bayesian inference (BI) analysis. The maximum parsimony (MP) analysis was conducted using PAUP* v4.0b1074. The MP searches included 1000 random addition replicates and TBR branch swapping using the MulTree option. ML analysis was performed using RaxML v. 8.0.575 with 1000 bootstrap replicates. BI analysis was carried out using MrBayes 3.2.276, with two independent runs of four simultaneous chains executed for 5,000,000 generations using the Markov chain Monte Carlo algorithm. Trees were sampled every 5000 generations, and the first 25% were discarded as burn-in. The trees were determined using a 50% majority-rule consensus to estimate PPs. Reconstructed trees were visualized using FigTree version 1.4.277.
Wild plants were collected and identified as Oyuntsetseg and Baasanmunkh. A voucher specimen was deposited in the herbarium of the National University of Mongolia (UBU) under voucher ID UBU0032866. Permission was obtained from the appropriate governing bodies. This study was conducted in accordance with the relevant guidelines and regulations.
The complete chloroplast genome of Adonis mongolica generated in this study was submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/) with GeneBank accession number OQ569932. All plastomes used in this study can be found in GenBank, and the GenBank accessions are shown in Additional File 1: Table S3. The other chloroplast genomes used in this study were downloaded from the NCBI.
Wang, WT Review of Adonis (Butterfly).Bull.Bot.Res.14 , 1–31 ( 1994 ).
Gao, T. et al. Evaluating the feasibility of using candidate DNA Barcodes in discriminating species of the large Asteraceae family. BMC Evol. Biol. 10, 324 (2016).
Conti, F. et al. Adonis fucensis (A. sect Adonanthe, Ranunculaceae), a new species from the Central Apennines (Italy). Biology (Basel) 12, 118 (2023).
Nishikawa, T. & Kadota, Y. Adonis L. In Flora of Japan (eds Iwatsuki, K. et al.) 287–288 (Kodansha Ltd, 2006).
Ren , Y. , Chang , H.-L. , Tian , X. , Song , P. & Endress , PK Floral development in Adonideae (Ranunculaceae).Flora Morphol.Distribution.Funct.Ecol.Plants 204, 506–517.
Son , DC , Park , BK & Ko , SC Phylogenetic study of the Adonanthe section of the genus Adonis L. (Ranunculaceae) based on ITS sequences.Korean J. Plant Taxon.46, 1–12 (2016).
Fu, D. & Robinson, O. R. Adonis. In Flora of China (eds Wu, Z. Y. et al.) 389–391 (Science Press; Missouri Botanical Garden Press, 2001).
Mitrenina, EY et al.Karyotype and genome size of Adonis amurensis and Adonis pennina (Ranunculaceae) from Asian Russia.Ukraine.J. Ecol.11, 163–170 (2021).
Mitrenina, E. Y. et al. Karyotypes and genome sizes of Adonis vernalis and Adonis volgensis. Turczaninowia 25, 5–15 (2022).
Basanmunkh, S. et al.Updated checklist of vascular plants endemic to Mongolia.Diversity 13, 619 (2021).
Basanmunkh, S. et al.Flora of Mongolia: An annotated checklist of native vascular plants.PhytoKeys 192, 63–169 (2022).
Article PubMed PubMed Central Google Scholar
Baasanmunkh , S. , Oyuntsetseg , B. & Tsegmed , Z. Distribution of vascular plants in Mongolia—Part IMr.J. Biol.Sci.Rev. 20, 3–28 (2022).
Hiroko, I.-N.et al.Assessment of pharmacological effects of Mongolian medicinal plant Adonis mongolica in guinea pigs in vivo and in vitro.Toho J. Med.3, 131–141 (2017).
Magsar , D. & Tsagaanmaam , D. Three cotyledons of Adonis mongolica Sim.Proc.Inst.Bot.Mr.Acad.Sci.10, 57–62 (1984).
Thieme, H. & Lamchav, A. Isolation of olitoriside and glucoolitoriside from Adonis mongolica Sim. Pharmazie 29, 610 (1974).
Dashzeweg, N., Batlai, O. & Radnaakhand, T. Mongolian Red List and Conservation Action Plans of PLANTS, vol. 9 (2011).
Heyn, C. C. & Pazy, B. The annual species of Adonis (Ranunculaceae)—A polyploid complex. Plant Syst. Evol. 168, 181–193 (1989).
Basanmunkh, S. et al.Distribution of vascular plants in Mongolia—Part II.Mr.J. Biol.Sci.20, 35–63 (2022).
Grubov, V. I. Key to the vascular plants of Mongolia (with an atlas), vols. I and II. Key to Vasc. plants Mong. 443 (1982).
Hoffmann, M. H. Ecogeographical differentiation patterns in Adonis sect. Consiligo (Ranunculaceae). Plant Syst. Evol. 211, 43–56 (1998).
Seidl, A. et al. Genotyping-by-sequencing reveals range expansion of Adonis vernalis (Ranunculaceae) from Southeastern Europe into the zonal Euro-Siberian steppe. Sci. Rep. 12, 1–14 (2022).
Zhang, S. D. et al. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367 (2017).
Article CAS PubMed Google Scholar
Li, H. T. et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470 (2019).
Ravi, V., Khurana, J. P., Tyagi, A. K. & Khurana, P. An update on chloroplast genomes. Plant Syst. Evol. 271, 101–122 (2008).
Johansson, J. T. There large inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae). Plant Syst. Evol. 218, 133–143 (1999).
Zhang, L. Q., Zhang, X. Y., Hu, Y. W., Sun, R. S. & Yu, J. L. Complete chloroplast genome of Adonis amurensis (Ranunculaceae), an important cardiac folk medicinal plant in east asia. Mitochondrial DNA Part B Resour. 6, 583–585 (2021).
He, J. et al. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci. Rep. 9, 1–13 (2019).
Zhai, W. et al. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogenet. Evol. 135, 12–21 (2019).
Article CAS PubMed Google Scholar
Kaneko , S. , Nakagoshi , N. & Isaki , Y. Origin of the endangered tetraploid Adonis ramosa (Ranunculaceae) assessed with chloroplast and nuclear DNA sequence data.Act Phytotaxon.Geobot.59, 165–174 (2008).
Suh, Y. et al. Molecular evidence for the taxonomic identity of Korean Adonis (Ranunculaceae). J. Plant Res. 115, 217–223 (2002).
Article CAS PubMed Google Scholar
Karahan, F., İlçim, A., Türkoğlu, A., İlhan, E. & Haliloğlu, K. Phylogenetic relationship among taxa in the genus Adonis L. collected from Türkiye based on nrDNA internal transcribed spacer (ITS) markers. Mol. Biol. Rep. 49, 7815–7826 (2022).
Hu, S. et al. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genom. 16, 1–14 (2015).
Asaf, S. et al. Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci. Rep. 10, 3881 (2020).
Article ADS CAS PubMed Google Scholar
Liu, H. et al. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front. Plant Sci. 9, 1–16 (2018).
Corbett-Detig, R. B. et al. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics 213, 1495–1511 (2019).
Article CAS PubMed PubMed Central Google Scholar
Charboneau, J. L. M., Cronn, R. C., Liston, A., Wojciechowski, M. F. & Sanderson, M. J. Plastome structural evolution and homoplastic inversions in Neo-Astragalus (Fabaceae). Genome Biol. Evol. 13, evab215 (2021).
Article CAS PubMed PubMed Central Google Scholar
Dong, W. et al. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genet. 15, 1–8 (2014).
Hoot, S. B. Phylogeny of the Ranunculaceae based on preliminary atpB, rbcL and 18S nuclear ribosomal DNA sequence data. Syst. Evol. Ranunculiflorae 251, 241–251 (1995).
Kim , YK , Park , CW & Kim , KJ Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia , and its evolutionary implications .Mol.Cells 27, 365–381.
Article CAS PubMed Google Scholar
Drescher, A., Ruf, S., Calsa, T. Jr., Carrer, H. & Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22, 97–104 (2000).
Article CAS PubMed Google Scholar
Park, I. et al. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One 12, 1–18 (2017).
Choi, J. E., Kim, G. B., Lim, C. E., Yu, H. J. & Mun, J. H. The complete chloroplast genome of Aconitum austrokoreense Koidz. (Ranunculaceae), an endangered endemic species in Korea. Mitochondrial DNA Part B Resour. 1, 688–689 (2016).
Subramanian, A. R. Molecular genetics of chloroplast ribosomal proteins. Trends Biochem. Sci. 18, 177–181 (1993).
Yu, B., Zhang, R., Yang, Q., Xu, B. & Liu, Z. L. The complete chloroplast genomes of Trollius farreri and Anemone taipaiensis (Ranunculaceae). Mitochondrial DNA Part B Resour. 4, 3270–3271 (2019).
Park, S., Jansen, R. K. & Park, S. J. Complete plastome sequence of (Ranunculaceae) and transfer of the gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15, 1–12 (2015).
Alqahtani, A. A. & Jansen, R. K. The evolutionary fate of rpl32 and rps16 losses in the Euphorbia schimperi (Euphorbiaceae) plastome. Sci. Rep. 11, 1–12 (2021).
Cusack, B. P. & Wolfe, K. H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 23, 270–272 (2007).
Article CAS PubMed Google Scholar
Ueda, M. et al. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402, 51–56 (2007).
Article CAS PubMed Google Scholar
Son, D. C., Park, B. K., Chang, K. S., Choi, K. & Shin, C. H. Cladistic analysis of the section Adonanthe under genus Adonis L. (Ranunculaceae) from East Asia. J. Asia-Pac. Biodivers. 10, 232–236 (2017).
Clegg, M. T. & Durbin, M. L. Flower color variation: A model for the experimental study of evolution. Proc. Natl. Acad. Sci. USA 97, 7016–7023 (2000).
Article ADS CAS PubMed PubMed Central Google Scholar
Durbin, M. L., Lundy, K. E., Morrell, P. L., Torres-Martinez, C. L. & Clegg, M. T. Genes that determine flower color: The role of regulatory changes in the evolution of phenotypic adaptations. Mol. Phylogenet. Evol. 29, 507–518 (2003).
Article CAS PubMed Google Scholar
Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Article CAS PubMed Google Scholar
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [(accessed on 28 March 2022)]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Babraham Institute, 2010).
Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 (2017).
Kurtz, S. et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642 (2001).
Article CAS PubMed PubMed Central Google Scholar
Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585 (2017).
Article CAS PubMed PubMed Central Google Scholar
Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Article CAS PubMed PubMed Central Google Scholar
Tillich, M. et al. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11 (2017).
Article CAS PubMed PubMed Central Google Scholar
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Article CAS PubMed Google Scholar
Lohse, M., Drechsel, O. & Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274 (2007).
Article CAS PubMed Google Scholar
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Article CAS PubMed PubMed Central Google Scholar
Babicki, S. et al. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153 (2016).
Article CAS PubMed PubMed Central Google Scholar
Ankenbrand, M. J., Hohlfeld, S., Hackl, T. & Förster, F. AliTV-interactive visualization of whole genome comparisons. PeerJ Comput. Sci. 3, e116 (2017).
Darling, A. E., Mau, B. & Perna, N. T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5, e11147 (2010).
Article ADS PubMed PubMed Central Google Scholar
Amiryousefi, A., Hyvönen, J. & Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031 (2018).
Article CAS PubMed Google Scholar
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, 273–279 (2004).
Brudno, M. et al. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 19, 54–62 (2003).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Article CAS PubMed Google Scholar
Addinsoft. XLSTAT Statistical and Data Analysis Solution (Addinsoft, 2021).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066 (2002).
Article CAS PubMed PubMed Central Google Scholar
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007).
Article CAS PubMed Google Scholar
Darriba, D., Taboada, GL, Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing.Nat. Methods 9, 772 (2012).
Article CAS PubMed PubMed Central Google Scholar
Swofford, D. L. & Documentation, B. Phylogenetic analysis using parsimony. Illinois Nat. Hist. Surv. Champaign (1989).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Article CAS PubMed PubMed Central Google Scholar
Ronquist, F. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Article PubMed PubMed Central Google Scholar
Rambaut, A. FigTree v1. 4. Molecular evolution, phylogenetics and epidemiology. Edinburgh Univ. Edinburgh, Inst. Evol. Biol. (2012).
This study was financed by a research project (a study on the distribution of vascular plants in Mongolia; Grant KNA1-2-38, 20-5) of the Korea National Arboretum, Republic of Korea, and the State Assignment of the CSBG SB RAS (Grant AAAA-A21-121011290024-5). This study was funded by the Financial Program on Basic Protection Science, 2023. This research was supported by Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2023R1A6C101B022). All the funders mentioned provided financial support for our study. We thank Ms. Ch. Javzandolgor, who provided excellent photographs of Adonis mongolica used in this study. This study was part of the first author’s PhD thesis.
Department of Biology and Chemistry, Changwon National University, Changwon, 51140, South Korea
Nudkhuu Nyamgerel, Shukherdorj Baasanmunkh, Inkyu Park & Hyeok Jae Choi
Department of Biology, School of Arts and Science, National University of Mongolia, Ulaanbaatar, 14201, Mongolia
Batlai Oyuntsetseg & Gun-Aajav Bayarmaa
Central Siberian Botanical Garden, Siberian Branch of the Russian Academy of Science, Novosibirsk, 630090, Russia
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
All authors contributed to the experimental design oversight. N.N. and I.P. analyzed the data. N.N., S.B., and I.P. wrote the draft. H.J.C., B.O., G.-A.B., and A.E. interpretated the data and revised the manuscript. H.J.C. and I.P. supervised the project. All authors read and approved the manuscript.
Correspondence to Inkyu Park or Hyeok Jae Choi.
The authors declare no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Nyamgerel , N. , Baasanmunkh , S. , Oyuntsetseg , B. et al.Insight into chloroplast genome structural variation in the tribe Adonideae.Sci Rep 13, 22014 (2023).https://doi.org/10.1038/s41598-023-49381-x
DOI: https://doi.org/10.1038/s41598-023-49381-x
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Scientific Reports (Sci Rep) ISSN 2045-2322 (online)

Magnetic Beads For Dna Purification Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.